A Student Workshop at Royal College of Art 2023
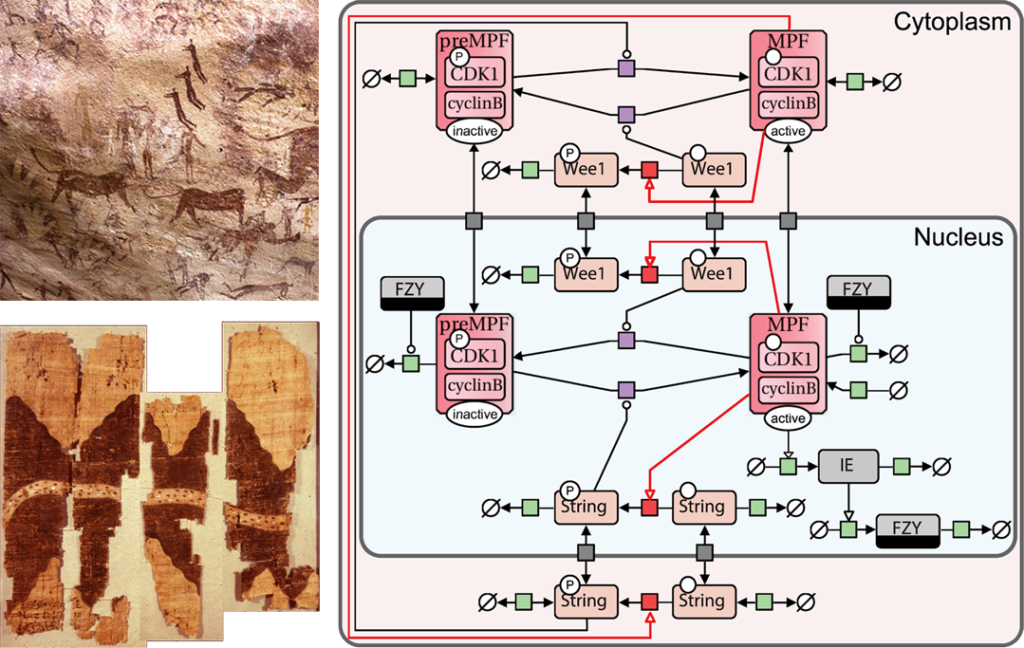
Since the era of our cave-drawing ancestors, humans have possessed innate cognitive faculties that are naturally engaged by diagrams. Diagrams notably play a key role in conveying information in biology, where they are often used to graphically represent the series of events taking place during a biological process. Such diagrams can be found in every good handbook for biology, but also in biology research papers, where they may be used to summarize the authors’ findings, for example. While these diagrams are key for the understanding of the biological processes they represent, they have for a long time remained inconsistent and ambiguous, due to a lack of a common notation. This became a major issue at the beginning of the 2000’s with the advent of computational systems biology, that focused on building, exchanging and analysing large and complex diagrams representing the molecular events occurring in whole biological systems. In other areas of science, standard visual languages such as Circuit diagrams (physics) or the Unified Modelling Language (computer science) had been proven useful to draw consistent and unambiguous diagrams, and to facilitate the use of software tools to do so.

Taking example on these standards, a community composed of biochemists, modellers, and computer scientists developed, from the beginning of the 2000’s, the Systems Biology Graphical Notation (SBGN), a set of standard visual languages designed for the representation of biochemical networks in a consistent and clear manner. SBGN promotes effective and accurate depiction, storage, visualization, exchange, and reusability of various types of biochemical networks, including gene regulation, metabolism, and cellular signalling networks. The notation consists of comprehensive sets of symbols that possess precise semantics, along with detailed syntactic rules that define their application and interpretation. SBGN is still actively developed by an open community, and has become the de facto standard for the representation of biochemical networks.
The Workshop
In 2022, we kicked-off the DESIGN X BIOINFORMATICS initiative with a workshop discussing several directions on how the design and bioinformatics community can join forces to come up with new ideas. There was a broad interest from your side regarding the different topics discussed, but also we were only able to give a quick preview into the different areas.
This year we want to kick-off a number of follow-up meetings with a workshop focusing on biological network visualization – exploring different opportunities on how to visualize and present interactions in biology. We will focus for this purpose on an existing standard: SBGN PD language.
During the workshop, you will learn the basics of network biology, get an introduction to SBGN, and we will introduce you to a tool which you can use to draw SBGN maps.
The design workshop will explore together with you different ideas how to improve network biology representations and visualizations.
We will come up with three different briefs and you will afterwards work in groups on this brief.
You will get familiar with network biology and will get a hands-on introduction to a tool which you can use to draw biological networks based on a well-established standard. The developers will be present at the workshop.
Participants will get a certificate for their participation.
Please note that this is an in-person workshop, there will be no online session running in parallel!
Student Competition
Moreover, we will announce a student competition during the workshop. Eligible for participation will be only those groups attending the workshop. You will have time until Sunday 18th June to submit your group project. The Winning Team will get 250 GBP and will of course be announced here on the website.
Workshop Activity Session
This interactive session will trigger the discussion between designers and bioinformaticians to explore future steps forward. This session will be organized by designers to provide insights how a typical design and ideation process would address these underlying questions.
The interactive session will happen hybrid – activities will be delivered online as well as in-person at the RCA. The presentation of the results will happen then online during a Zoom session.
Time
Friday, June 2 2023, 9:30 am to 4:30 pm.
Registration
RCA students can register here until May 21 midnight.
Location
Royal College of Art
Studio Building
Howie Street
London SW11 4AY
United Kingdom
https://goo.gl/maps/zBU6rgP5qAToomyn9
Image References
Clemens Schmillen, 2014: Cave of the Beasts – cut out. Creative Commons Attribution-Share Alike 3.0 Unported.
Zyzzy – Photograph at the Turin Museum courtesy of J. Harrell, 2005: Fragments of Turin papyrus. Public Domain.
Vasundra Touré et al, 2018: PD map of the Drosophila cell cycle. Creative Commons Attribution License.

